Forces and elasticity in cell adhesion
 |
 |
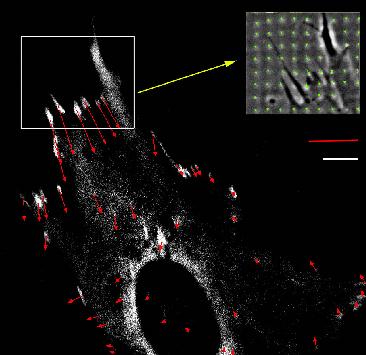
| Calculation of forces at focal adhesions from elastic substrate
data. Forces (red) exerted by an adhering fibroblast at sites of focal
adhesions (white) which are fluorescently marked by GFP-vinculin. The inset
shows a phase contrast image of the deformation of the micro-patterned
elastic substrate (green), from which the force pattern has been calculated.
Balaban et al., Nature Cell Biology 2001. |
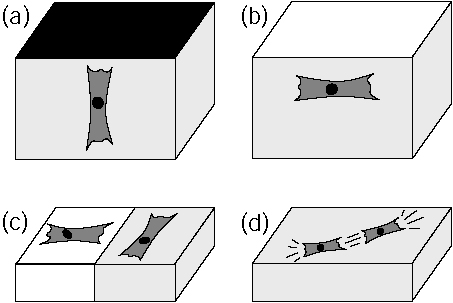
Predicted cell organization in soft media due to active mechanosensing.
In some situations, mechanical input signals can be as important for cellular
decision making as biochemical input signals. Cells actively sense the
elastic properties of their environment and strengthen contacts and cytoskeleton
in the direction of large effective stiffness. Therefore they orient (a)
perpendicular to clamped boundaries, (b) parallel to free boundaries, (c)
perpendicular and parallel on the soft and stiff sides of a rigidity gradient,
respectively, and (d) in parallel in and on a homogeneous medium.
Bischofs and Schwarz, PNAS 2003. |
Force is ubiquitous in biological systems. Many cellular programs
like cell division, cell adhesion and cell locomotion involve the
generation of physical force. Just imagine that there are many cells
which move inside you while you read this text: eg leukocytes circle
your body using the blood flow on their search for sites of
inflammation; rolling adhesion on the vessel walls allows them to
survey it for signs of inflammation, and has to function under large
shear forces; on encountering certain signals, leukocytes leave the
blood stream by squeezing through the vessel wall; in the surrounding
tissue, they crawl by pulling themselves along fibers. Or think of
fibroblasts which structure the connective tissue (which fills the
space between organs and tissues in the body) and close your wounds by
strongly pulling on their surrouding. More examples for cellular
forces and cell movements come from embryogenesis, morphogenesis, and
from malignent tumors, when cells do not respond to inhibiting signals
to crawling.
For cells adhering to flat rigid surfaces, cellular force is
generated mainly by actomyosin contractility and transmitted to the
substrate through sites of so-called focal adhesion. Focal adhesions
have both structural and signalling functions. From a structural point
of view, they provide the pinning sites needed by the cell in order to
spread out on the surface. From a signalling point of view, focal
adhesions trigger different signalling pathways which signal
successful adhesion to the cell; lack of these signals can in fact
lead to programmed cell death. During my postdoctoral work at the
Weizmann Institute, I became involved in a collaboration with
experimentalists in the Department of Cell Biology which succeeded in
measuring cellular forces at the level of focal adhesions for the
first time (Nature Cell Biology 2001, Biophysical Journal 2002). The
experimental technique combined the use of microstructured elastic
substrates and fluorescense marking of focal adhesions proteins. The
theoretical part consisted in calculating the force pattern from the
displacement pattern, which numerically amounts to solving an
ill-posed inverse problem. We were able to show that size and
direction of forces acting at focal adhesions correlate strongly with
size and elongation of the adhesion plaques as monitored by
GFP-vinculin (for fibroblasts, typical forces at focal adhesions are
10 nN, with a stress constant around 5.5 nN / micron^2). Thus force
and protein assembly (which in turn correlates with signalling) are
intimately linked at focal adhesions. However, the details of the
molecular mechanisms which cause the correlations between force,
protein assembly and signalling at focal adhesions are still elusive.
Recently we have improved the workflow required to measure cellular
traction patterns. Our improvements in traction force microscopy
include simultaneous use of several kinds of fluorescent markers,
advances in image processing and filtering, and flexible use of
different force reconstruction algorithms, eventually leading to a
spatial resolution of one micrometer (Sabass et al., Biophysical
Journal 2008). We then applied this technique to correlate for the
first time traction force with retrograde flow, which is a crucial
element in cell migration and can be measured with speckle
fluorescence microscopy. Combining it with traction force microscopy
allowed us to identify a biphasic relation between flow and force
(Gardel et al., Journal of Cell Biology 2008). In detail, we found
that actin speed is inversely related to traction stress near the cell
edge, indicating that the flow is increasingly impeded as the actin
becomes increasingly engaged to the adhesions. In contrast, larger FAs
where the actin speed is low are marked by a direct relationship
between actin speed and traction stress. The crossover between these
two regimes occurs at at threshold flow of around 10 nm/s, independent
of many possible determinants like actin polymerization or myosin
activity. Thus it appears that actin speed is a fundamental regulator
of traction force at FAs during cell migration.
Continuing our collaboration with the Gardel lab, recently we were
able to demonstrate that the measurement of cell-matrix forces can
also be used to infer cell-cell forces (Maruthamuthu et al., PNAS
2011). In particular, we found that cell-cell forces have a similar
magnitude as do cell-matrix forces, and that they increase with
increasing substrate stiffness. Our results suggest that cells balance
a similar amount of tension with their environment both as single
cells on a substrate and as part of a cell sheet.
In a collaboration with Julien Colombelli and Ernst Stelzer from
the EMBL at Heidelberg, we were able to show that force also
correlates with protein localization in stress fibers (Colombelli et
al., Journal of Cell Science 2009). Using laser cutting, we were
able to show that stress fibers are attached along their length to
the substrate. Upon cutting, these crosslinks restrict the length
over which the fiber can retract. In addition, the crosslinks become
tensed and then recruit zyxin, a protein which is known to be strongly
mechanosensitive. Using a theoretical model for stress distribution
along stress fibers, we found that zyxin localization follows
exactly the spatial distribution of force along the fiber. Therefore
stress fibers act as spatially distributed mechanosensors.
In earlier work we had shown that also focal adhesions grow under
the application of external force, suggesting that focal adhesions
function as spatially localized mechanosensors (Riveline et al.,
Journal of Cell Biology 2001). Together with other experimental
studies, our results demonstrate that animal cells have evolved a
sophisticated apparatus which they can use to actively sense the
mechanical properties of the environment. In fact it has long been
known, especially in the medical and bioengineering communities, that
cell organization in soft media is strongly influenced by the
mechanical properties of the environment. Our experimental results
now suggest that this cell organization is resulting from active
processes on the level of single cells.
In order to predict and
control these cellular processes (for example for applications in
tissue engineering), it is essential to establish a theoretical
framework for the interplay of mechanical activity of cells and
mechanical properties of the surrounding matrix. We have shown
that a large body of experimental observations on cell organization in
soft media can be consistently explained from a relatively simple
theory which describes active cell behavior by an extremum principle
in linear elasticity theory (PNAS 2003, Physical Review E 2004,
Physical Review Letters 2005). Our theory uses concepts which have
been developed in the 70s for elastic interactions of atomic defects
in crystals. In detail, we model cells as anisotropic force
contraction dipoles and solve the elastic equations for elastic
isotropic material with different geometries and boundary conditions
(Physical Review Letters 2002). We start from the observation that
cells strengthen contacts and cytoskeleton in the direction of large
effective stiffness in their environment (possibly because build-up of
force at focal adhesions is more efficient in a stiff environment) and
show that this corresponds to a minimization of the quantity W =
uij Pij, where uij is the strain
tensor of the surrounding medium (including image strain in the case
of finitely sized geometries) and Pij is the force dipole
tensor representing the cell. From this principle, we show that cells
orient in the direction of external tensile strain, that they orient
parallel and normal to free and clamped surfaces, respectively, and
that they interact elastically to form strings, in excellent agreement
with experimental observations. We also extended this
framework to deal with matrix-mediated structure formation in
ensembles of cells (Acta Biomaterialia 2006).
Publications from collaborations with experimentalists:
-
N. Q. Balaban, U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur,
I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi and
B. Geiger, Force and focal adhesion assembly: a close relationship
studied using elastic micro-patterned substrates, Nat. Cell
Biol. 3: 466-472 (2001) (abstract, PDF)
-
D. Riveline, E. Zamir, N. Q. Balaban, U. S. Schwarz, B. Geiger,
Z. Kam, A. D. Bershadsky, Focal contact as a mechanosensor: externally
applied local mechanical force induces growth of focal contacts by a
mDia1-dependent and ROCK-independent mechanism, J. Cell
Biol.153: 1175-1185 (2001) (abstract, PDF)
-
U. S. Schwarz, N. Q. Balaban, D. Riveline, A. Bershadsky, B. Geiger,
S. A. Safran, Calculation of forces at focal adhesions from elastic
substrate data: the effect of localized force and the need for
regularization, Biophys. J. 83: 1380-1394 (2002) (abstract, PDF)
-
U. S. Schwarz, N. Q. Balaban, D. Riveline, L. Addadi, A. Bershadsky,
S. A. Safran, B. Geiger, Measurement of cellular forces at focal
adhesions using elastic micro-patterned substrates,
Mat. Sci. Eng. C 23: 387-394 (2003) (abstract, PDF)
-
C. Cesa, N. Kirchgessner, D. Meyer, U.S. Schwarz, B. Hoffmann and R. Merkel.
Micropatterend silicone elastomer substrates for high resolution analysis of cellular force patterns.
Rev. Sci. Instruments, 78:034301, 2007.
(abstract,
doi:10.1063/1.2712870,
PDF)
-
B. Sabass, M. L. Gardel, C. Waterman and U. S. Schwarz.
High resolution traction force microscopy based on experimental and computational advances.
Biophys. J., 94:207-220, 2008.
(abstract,
doi:10.1529/biophysj.107.113670,
PDF)
-
M. L. Gardel, B. Sabass, L. Ji, G. Danuser, U. S. Schwarz, and C. M. Waterman.
Traction stress in focal adhesions correlates biphasically with actin
retrograde flow speed. J. Cell Biol., 183:999-1005, 2008.
(abstract,
doi:10.1083/jcb.200810060,
PDF)
- Venkat Maruthamuthu, Benedikt Sabass, Ulrich S. Schwarz, and Margaret L. Gardel.
Cell-ECM traction force modulates endogenous tension at cell-cell contacts.
Proc. Natl. Acad. Sci. USA, 108:4708-4713, 2011.
(abstract,
doi:10.1073/pnas.1011123108,
PDF, Supplemental material)
-
J. Colombelli, A. Besser, H. Kress, E.G. Reynaud, P. Girard, E. Caussinus,
U. Haselmann, J.V. Small, U. S. Schwarz, and E.H.K. Stelzer.
Mechanosensing in actin stress fibers revealed by a close correlation
between force and protein localization. J. Cell Sci., 2009.
(abstract,
doi: 10.1242/jcs.042986,
PDF)
Theoretical work on cell organization in soft media:
-
U. S. Schwarz and S. A. Safran, Elastic interactions of cells,
Phys. Rev. Lett. 88: 048102 (2002) (abstract, cond-mat/0201110, PDF)
-
I. B. Bischofs and U. S. Schwarz, Cell organization in soft media due
to active mechanosensing, Proc. Natl. Acad. Sci. USA
100: 9274-9279 (2003) (abstract, cond-mat/0308187,
PDF)
-
I. B. Bischofs, S. A. Safran and U. S. Schwarz, Elastic interactions
of active cells with soft materials, Phys. Rev. E 69: 021911 (2004)
(abstract,
cond-mat/0309427,
DOI: 10.1103/PhysRevE.69.021911
PDF)
-
S. A. Safran, N. Gov, A. Nicolas, U. S. Schwarz and T. Tlusty,
Physics of cell elasticity, shape, and adhesion,
Physica A 352: 171-201 (2005)
(abstract,
PDF)
-
I. B. Bischofs and U. S. Schwarz,
Effect of Poisson ratio on cellular structure formation,
Phys. Rev. Lett. 95: 068102 (2005)
(abstract,
cond-mat/0504097,
DOI: 10.1103/PhysRevLett.95.068102,
PDF)
-
U. S. Schwarz and I. B. Bischofs.
Physical determinants of cell organization in soft media.
Med. Eng. Phys., 27: 763-72 (2005)
(abstract,
PDF)
- U. S. Schwarz, T. Erdmann, and I. B. Bischofs.
Focal adhesions as mechanosensors: the two-spring model.
BioSystems, 83: 225-232 (2006)
(abstract,
q-bio.SC/0608006,
PDF)
- I. B. Bischofs and U. S. Schwarz.
Collective effects in cellular structure formation mediated by
compliant environments: a Monte Carlo study.
Acta Biomaterialia, 2: 253-265, 2006.
(abstract,
cond-mat/0510391,
doi:10.1016/j.actbio.2006.01.002,
PDF)
Last modified Di 4. Okt 17:45:17 CEST 2011
by USS.
Back to home page Ulrich Schwarz.

